MAJOR AREAS OF CURRENT RESEARCH:
In order to achieve our overall scientific career goal of understanding how small RNAs
and Transcription Factors work together in living cells, our research objectives for the
current time frame are (a) to identify the function of specific circuits by studying their
dynamic behavior in vivo using Arabidopsis thaliana as a model organism, and (b) to
apply the computational and experimental tools developed in order to compare the
structure and function of TF-miRNA circuits within Arabidopsis and across plant
species. Our research program currently has two main project tracks in pursuing these
goals:
(1) Regulatory Genomics: Comprehensive mapping of transcriptional control regions.
The lack of precise data on the location of transcriptionally active regions in plant
genomes currently hampers large-scale identification of functional cis-regulatory control
elements. To address this barrier, the experimental portion of this track focuses on
mapping of transcriptional control regions using deep sequencing and open chromatin
assays. One project underway is transcription start site (TSS) identification for
microRNAs and protein-coding genes in Arabidopsis root samples using nanoCAGE-
2000, a method we have developed in the laboratory which sequences tags attached to
the 5' caps of gene transcripts using a relatively small amount of total RNA. This
method is important for promoter analysis of tissue-specific and cell-type specific
samples. The computational projects within this track include the analysis of TSS
distribution shapes, and identification of enrichment regions for proximal promoter
elements. A second phase of this project now underway is to overlay probable regions
of open chromatin to obtain a set of high-confidence transcription factor binding site
predictions upstream of TFs, other protein-coding genes, and miRNAs.
(2) Network Analysis: Identification of TF-miRNA gene circuits. Projects within this track
focus on circuit structure at three different levels of inquiry using computational
methods: network motif analysis, inference using functional genomics data, and
analysis of conserved structures. Our first project identifies over-represented network
substructures (“network motifs”) among the three different types of relevant nodes (TFs,
miRNAs, and non-TF genes) by using and modifying existing approaches to deal with
the unique behaviors of each node type. A second project is underway to develop an
edge-weighted model which uses gene expression data to identify those over-
represented structures which are most likely to function within a specific plant cell type.
This project begins by building a Bayes Net on small acyclic structures, and then
identifies the best scoring model according to an appropriate metric. A more involved
goal is to extend this idea to analysis of networks containing directed cycles.


Arabidopsis
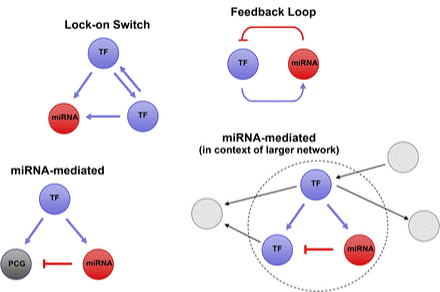
miRNA-containing regulatory circuit examples
pScarecrow-NTF-GFP-Root-endodermis-specific-synthetic-construct
Protoplast Expresssing SPL14 Driven GFP

